Projects
Here are some examples of the projects carried out in the Lucky Lab!
Featured

SPACETIME is an EU consortium studying how spatial cellular organization changes with disease severity in non-small cell lung carcinoma (NSCLC). We are collaborating with biologists, clinicians, biotechnologists, and other computational biologists or machine learning scientists to build a spatial multiomic map of NSCLC across species and disease stages with the goal of building better diagnostics, expanding our understanding of disease mechanisms, and developing treatment options to improve outcomes for lung cancer patients.
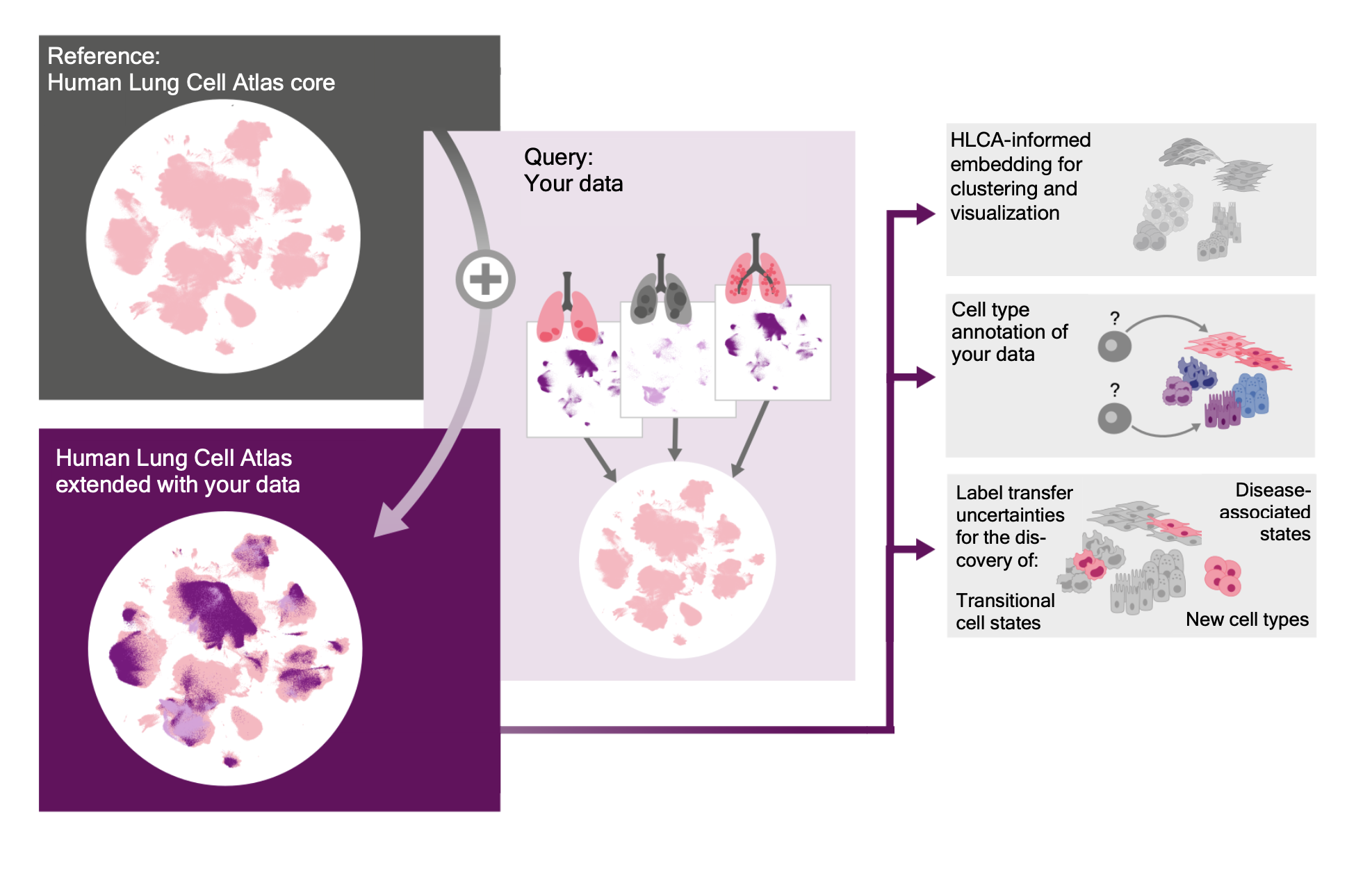
The integrated Human Lung Cell Atlas (HLCA) represents the first large-scale, integrated single-cell reference atlas of the human lung. It consists of over 2 million cells from the respiratory tract of 486 individuals and includes 49 different datasets. The HLCA is widely used in the community, with over 2,000 downloads of the model alone.
We are participating in an EU-funded doctoral network on understanding mechanisms, identifying biomarkers, and developing treatments for chronic lung diseases including Asthma and COPD. We interact with groups across disciplines to build tools and analyse data to generate insights in asthma and COPD with translational impact.
More
Our goal is to provide an open source, community driven, extensible platform for continuously updated benchmarking of formalized tasks in single-cell analysis.

We are heavily involved in the Human Cell Atlas (HCA) and are in a leading role in the HCA integration team. Here we coordinate an international group of PhD students and postdocs to build the first version of the HCA, a cellular reference atlas for each human tissue or organ. In 17 projects we work together with the respective HCA organ bionetwork coordinators and their champions to integrate datasets of sufficient quality to build a consensus cellular representation of that organ. We work together with the HCA data repository, the CELLxGENE team (lattice), and the HCA cell annotation project (CAP) team to ingest data, and annotate the resulting objects to build a community consensus annotation. Examples of these projects include the Human Lung Cell Atlas v1, and the Human Retinal Cell Atlas.

We benchmarked 68 method and preprocessing combinations on 85 batches of gene expression, chromatin accessibility and simulation data from 23 publications, altogether representing >1.2 million cells distributed in 13 atlas-level integration tasks. We evaluated methods according to scalability, usability and their ability to remove batch effects while retaining biological variation using 14 evaluation metrics.
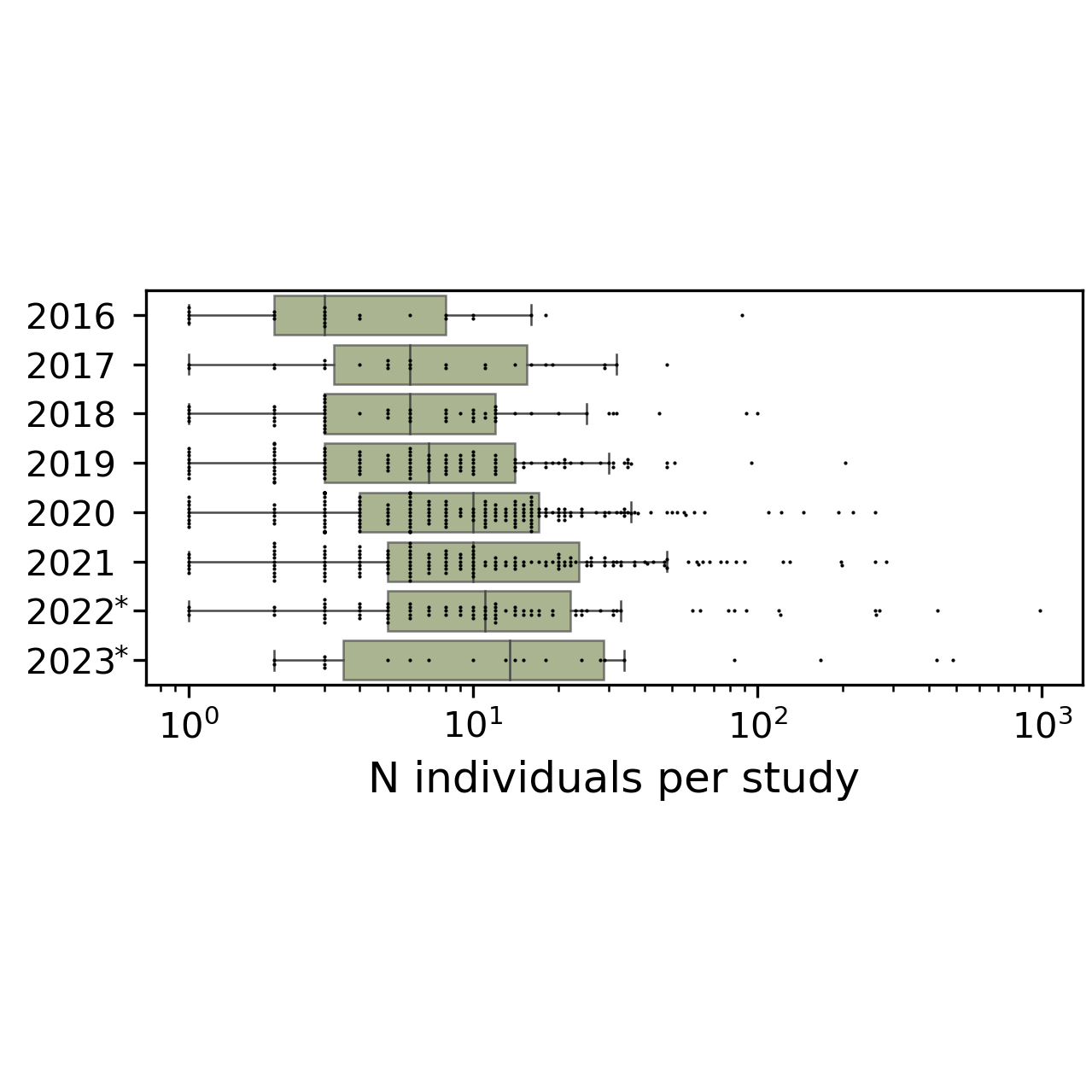
Single-cell datasets are growing in size, and slowly also in the number of samples profiled. Additionally, integrated reference atlases that contain hundreds to thousands of samples are becoming increasingly available. This enables studying inter-individual variation from single-cell datasets. We have pioneered the study of inter-individual variation on single-cell data by aggregating datasets (Muus et al., Nat Med 2021), extended this to understand the effects of demographic variation on human lung transcriptomes (Sikkema et al., Nat Med 2023), and are developing and benchmarking approaches that represent inter-sample variation. These approaches have the potential for diagnostic and prognostic impact by taking a wholistic view of the cells from a tissue, rather than inspecting each cell or cell type individually.